Scientists from CIIRC CTU and IOCB Prague lead a benchmarking effort for AI-driven discovery of molecules

In April 2024, brothers Roman and Anton Bushuiev from the teams of Tomáš Pluskal at IOCB Prague and Josef Šivic at CIIRC CTU initiated a collaboration between experts from 14 research institutes across the globe to benchmark AI methods for the discovery of molecules from mass spectrometry data. The collaborative project, titled MassSpecGym, aims to spark the development of next-generation machine learning models for identifying new molecules from nature with applications spanning drug development, environmental science, or space exploration.
The first success didn’t take long to come. The results of the cross-disciplinary initiative were already presented as a Spotlight poster at one of the world’s top machine learning conferences – NeurIPS 2024 in Vancouver, in December 2024.
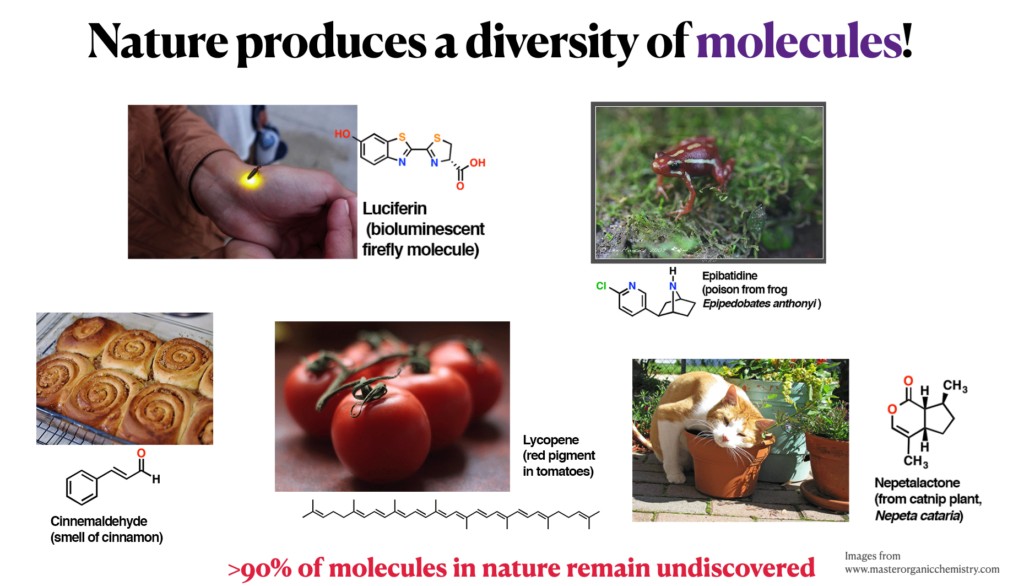
The discovery of small molecules profoundly influences numerous scientific fields such as organic chemistry, molecular biology, drug development, and environmental analysis. Despite advancements, only a small fraction of life’s molecular diversity has been uncovered.
Tandem mass spectrometry (MS/MS) is a cornerstone instrumental technique for identifying molecular structures from biological and environmental samples, enabling applications such as discovering bioactive compounds for drug development, optimizing drug dosages in clinical settings, and detecting environmental pollutants at trace levels. At its core, a tandem mass spectrometer fragments molecules and records the masses of these fragments in so-called MS/MS spectra.
“A typical biological or environmental sample produces thousands of tandem mass spectra, each representing a distinct molecule. Yet, annotating these spectra with molecular structures remains a challenge, with fewer than 10% of spectra successfully annotated using state-of-the-art machine learning methods. This leaves much of the chemical space uncovered, limiting our ability to unlock new scientific and technological advancements,” says Tomáš Pluskal from IOCB Prague.
Currently, the development of new AI methods for mass spectrometry is limited by the absence of well-standardized training datasets and evaluation protocols. The project “MassSpecGym: A benchmark for the discovery and identification of molecules” addresses this limitation.
“Machine learning benchmarks such as ImageNet revolutionized the field of AI by standardizing development, evaluation, and assessment of progress. Similarly, we propose a benchmark for molecular discovery to tackle the critical challenge of annotating tandem mass spectra and aim to foster a new generation of AI models for uncovering the undiscovered space of chemical structures present in nature,” explains doctoral student and the main author of the project Roman Bushuiev.

MassSpecGym comprises three core components: (i) the largest publicly available dataset of tandem mass spectra labeled with molecular structures, (ii) three well-defined machine-learning challenges rendering the process of molecular discovery from mass spectra into well-defined computational problems, and (iii) carefully-selected held-out pairs of mass spectra and molecules designed to evaluate the ability of AI models to generalize to new chemical space. Additionally, MassSpecGym provides a user-friendly platform for developing and evaluating new AI models.
A research paper on MassSpecGym was selected for a Spotlight poster presentation at NeurIPS 2024 in Vancouver, which is one of the most prestigious conferences in machine learning and is ranked among the top ten publication venues in all areas of science by Google Scholar.
This research was co-funded by EU projects FRONTIER (No. 101097822) and ELIAS (No. 101120237).
Original article: R. Bushuiev, A. Bushuiev, N. F. de Jonge, A. Young, F. Kretschmer, R. Samusevich, J. Heirman, F. Wang, L. Zhang, K. Dührkop, M. Ludwig, N. A. Haupt, A. Kalia, C. Brungs, R. Schmid, R. Greiner, B. Wang, D. S. Wishart, L.-P. Liu, J. Rousu, W. Bittremieux, H. Rost, T. D. Mak, S. Hassoun, F. Huber, J. J. J. van der Hooft, M. A. Stravs, S. Böcker, J. Sivic, T. Pluskal, “MassSpecGym: A benchmark for the discovery and identification of molecules”, Advances in Neural Information Processing Systems (NeurIPS), 2024. https://doi.org/10.48550/arXiv.2410.23326






