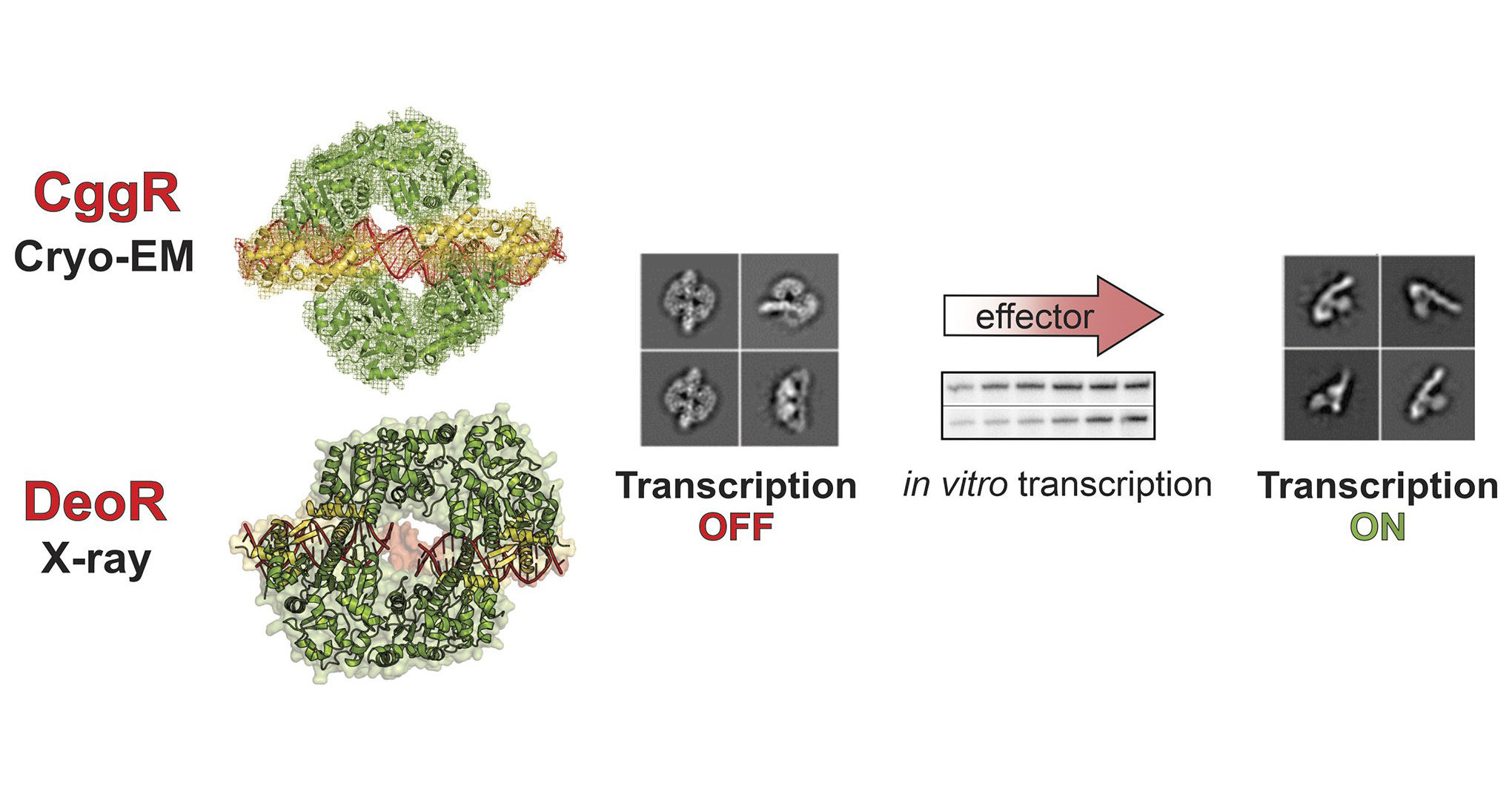
Researchers have provided new insights into bacterial gene regulation in a study published in Nucleic Acids Research. Transcriptional regulators, proteins that act as molecular switches turning on and off the expression of genes, are crucial for bacteria to adapt to environmental changes. The proteins of the large SorC family play a crucial role in controlling carbohydrate metabolism and quorum sensing. The study focuses on two such regulators, DeoR and CggR from Bacillus subtilis, each representing repressors of one SorC subfamily.
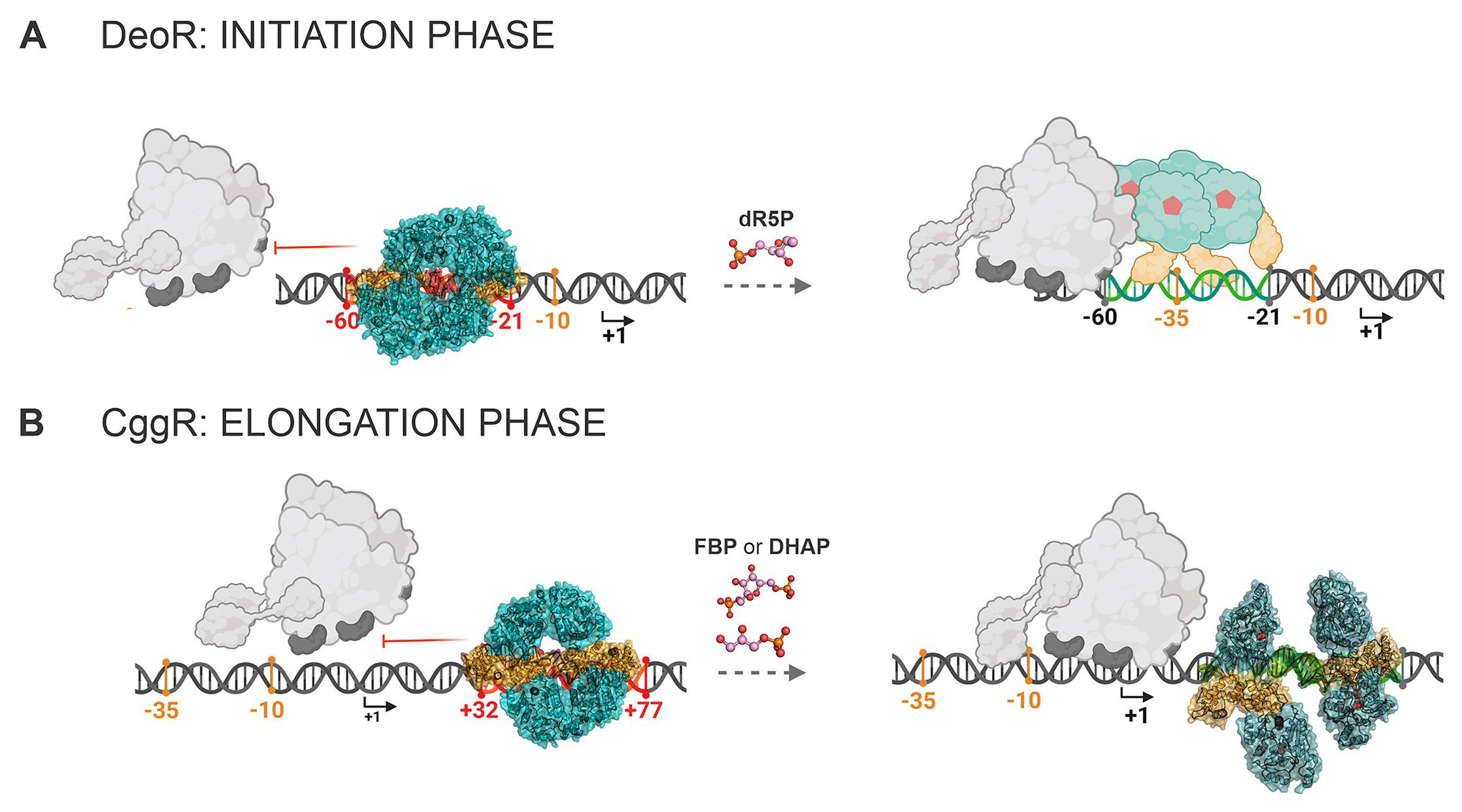
The research, led by Pavlína Řezáčová from IOCB Prague, with Markéta Šoltysová as the first author, utilized X-ray crystallography and cryo-electron microscopy to uncover how these proteins interact with DNA. They discovered that despite both DeoR and CggR forming tetrameric structures when bound to DNA, their mechanisms of action differ. While CggR contacts its DNA target in a highly cooperative dimer-by-dimer manner, DeoR, on the other hand, remains in the tetrameric form for its entire life cycle.
Moreover, the study identified both fructose-1,6-bisphosphate (FBP) and dihydroxyacetone phosphate (DHAP) as effectors for CggR. These molecules bind to CggR and cause it to release its hold on the gapA operon, demonstrating a broader spectrum of effector specificity most probably characteristic of SorC/CggR proteins.
This study provides valuable insights into the structural and functional mechanisms of SorC family repressors, advancing our knowledge in bacterial gene regulation. Investigating other SorC family members in different bacterial species could reveal whether these mechanisms are conserved. Additionally, understanding these regulatory mechanisms could aid in designing custom regulatory circuits for industrial and medical uses.
Read the paper:
- Šoltysová, M.; Škerlová, J.; Pachl, P.; Škubník, K.; Fábry, M.; Sieglová, I.; Farolfi, M.; Grishkovskaya, I.; Babiak, M.; Nováček, J.; Krásný, L.; Řezáčová, P. Structural characterization of two prototypical repressors of SorC family reveals tetrameric assemblies on DNA and mechanism of function. Nucleic Acids Res. 2024, gkae434. https://doi.org/10.1093/nar/gkae434